Monitoring rAAV Capsid Stability and Genome Ejection Using ARGEN

Introduction
Recombinant adenoviral vectors (rAAVs) have delivered robust treatment outcomes for a variety of monogenic disorders. They encode and deliver gene(s) to the host cell with the intent to mitigate or cure disease conditions. The canonical rAAV vector contains ≦ 4.7 kb, single-stranded (ss) DNA within a 60 protein icosahedral capsid, and tropism is selected from thirteen known natural and additional novel hybrid serotypes permitting precise tissue targeting (1,2).
The successful development of efficacious rAAV vectors can be challenging and requires a comprehensive understanding of the stability landscape under a range of thermal, chemical, storage, and mechanical (stirring) stress conditions. Quantitative and qualitative stability data permits the design of stabilizing solution condition(s), as well as the characterization of destabilizing effects resulting from purification, processing, manufacturing, storage, and freeze-thaw cycling. The rAAV capsid stability experiments in this technical note were performed to investigate and monitor the solution behavior of full and empty rAAV capsids under thermal stress using the ARGEN instrument.
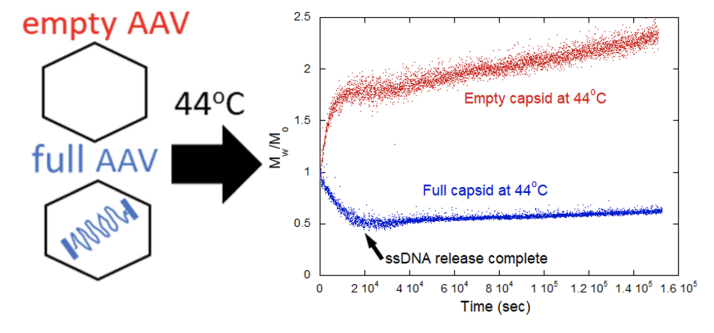
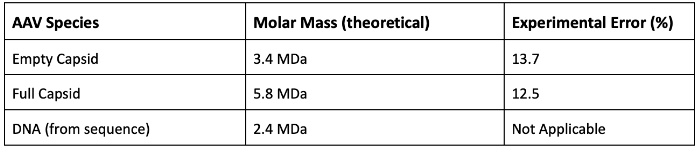
ARGEN is a benchtop lab instrument that enables a smart and rapid approach to therapeutic biopolymer development. The static light scattering platform addresses key challenges in biopharmaceutical R&D. ARGEN utilizes fixed angle (90°), SMSLS (simultaneous multiple sample light scattering) technology which provides rapid, real-time, continuous data collection for characterizing qualitative and quantitative properties of target molecules in situ. The device is equipped with multiple independently controlled sample cells, permitting the user to establish thermal, chemical, and mechanical (stirring) stress parameters on each sample concurrently. This allows for a highly flexible approach to experimental design. The ARGEN control software features an intuitive interface for all aspects of experimental design and independent control of each cell for parallel parameter adjustment and real-time data processing. rAAVs were produced by triple transfection in HEK-293 cells and purified from crude cell lysate by ion-exchange chromatography. Empty (without DNA) and full capsids (with 4.7 kb, ssDNA) were concentrated and dialyzed into 20mM sodium phosphate, 180mM NaCl at pH 7.3. Prior to the experiments, full and empty capsid samples were filtered using a 0.22µm cellulose acetate filter. Data was collected using ARGEN which is equipped with eight autonomous sample cells, permitting independent experimental parameter (temperature and stirring rate) adjustments. Additionally, each sample cell is engineered with a dedicated optical detector train. Absolute molecular weight (Mw) determination of full and empty capsids was achieved using the Zimm approximation. Mw(t) is the molecular weight (average) of the capsid (full or empty) as represented by changes in time resolved light scattering intensity, and Mo is the initial molecular weight of the capsid (full or empty) prior to aggregation at t = 0. Mw(t)/Mo represents the temporal change in molecular weight (average) with respect to the native state of the capsid(s). All experiments were performed in 1 cm pathlength, disposable cuvettes with 1 ml sample volumes. Characterizing viral rAAV capsid stability with or without DNA cargo and DNA ejection dynamics can provide insights into the role of DNA in stabilizing capsid architecture. For these experiments, ARGEN was used to define temporal changes in molecular weight (average) for the full and empty rAAV at 44°C (Figure 1). Empty capsids rapidly formed higher order aggregates due to colloidal instability. This provides possible evidence into the role of DNA in the maintenance of capsid architecture as evidenced by a rapid increase in molecular weight (average). Analysis of full capsids demonstrated the complete ejection of DNA in ~5.6 hours (~20000s) from the outset of the experiment. Following full evacuation of the genome, capsid protein aggregation proceeded slowly. This phenomena may occur due to the binding of capsid proteins to the ejected DNA, due to the large net negative charge, and subsequently preventing the formation of higher order protein species (aggregates). Further investigation is required to fully understand the differences in aggregation rates of empty and full capsid associated proteins. The successful development of an rAAV vector system requires quantitative and qualitative characterization of all vector components. Absolute molecular weights for the full and empty AAV capsids were determined using the Zimm approximation, neglecting the interaction term (A2) from data generated using ARGEN. Table 1 provides theoretical and experimental molar masses along with associated fractional errors. Fractional errors are due to concentration uncertainties (pooling of mixed and empty capsids), as well as neglect of the interaction term (A2) and Mie scattering effects (angular dependence of non-Rayleigh scatterers). ARGEN is uniquely suitable for in situ and real-time characterization of solution stability of larger species such as rAAVs and LNP-RNA complexes. This case study demonstrates ARGEN’s ability to elucidate differences in full and empty capsid stability and monitor genome ejection. This data is critical for optimizing rAAV formulations for stabilization during storage and manufacturing. Defining quantitative and qualitative properties of viral vector systems is critical to ensure maximum efficacy and stability during processing, storage, and parenteral administration. These experiments confirmed the ability of ARGEN to monitor DNA ejection, solution behavior, and the rAAV capsid stability of full and empty vectors under thermal stress, while also determining the absolute molecular weights of AAV vector and vector components. This data can provide a comprehensive stability landscape for the successful development of AAV vector systems, as well as LNP-RNA complexes. ARGEN™: Accelerating Biotherapeutic Formulation Development
How ARGEN™ Works
ARGEN™ Intuitive Control Software
Materials and Methods
Materials
Methods
Results
Monitoring rAAV Capsid Stability Behavior Under Isothermal Conditions
Molar Mass Determination of Full and Empty rAAV Capsids using ARGEN™
The Value of ARGEN™ in rAAV Capsid Stability Research
Conclusion
References